Mapping Drought Tolerance in Chickpea: Insights from Whole Genome Resequencing of a MAGIC Population
In our ongoing long-term collaboration with Dr. Rajeev Varshney, our team has taken a further step forward in understanding and enhancing drought tolerance in chickpea (Cicer arietinum). Our recent study, published in The Plant Genome, harnessed the power of whole genome resequencing (WGRS) and detailed phenotyping of a multi-parent advanced generation intercross (MAGIC) population to map key drought tolerance traits at a high resolution.
Key Findings
-
Extensive Genomic Resource:
We generated 4.67 TB of high-quality clean data from 52.02 billion Illumina reads obtained from a MAGIC population comprising 1136 recombinant inbred lines (RILs) and eight elite founder lines. This effort led to the identification of over 930,000 SNPs, along with tens of thousands of insertions and deletions. Using these variants, we constructed a high-density genetic map spanning 1606.69 centimorgans with 57,180 SNPs aggregated into 13,172 marker bins. -
High Resolution Mapping of Drought Tolerance:
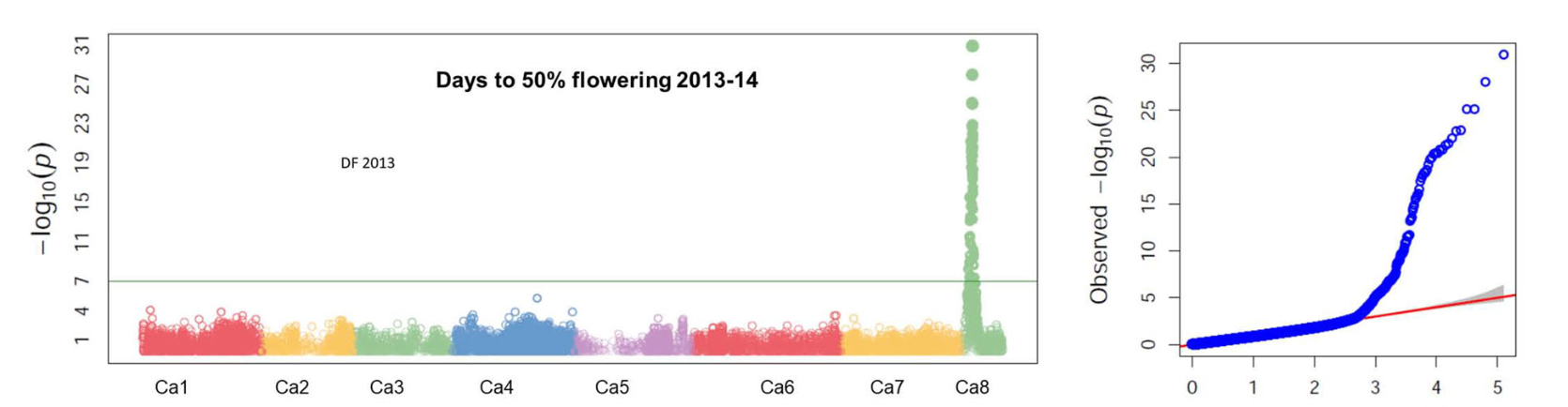
By employing a compressed mixed linear model for a genome-wide association study (GWAS), we detected 737 significant marker-trait associations (MTAs) for several agronomic traits related to drought tolerance—including days to 50% flowering, days to maturity, plant height, 100 seed weight, biomass, and harvest index. Moreover, a complementary identity-by-descent (IBD)-based mixed model approach uncovered additional quantitative trait loci (QTLs), with some major QTLs explaining high proportions of the observed phenotypic variation. -
Candidate Genes with Breeding Potential:
Key candidate genes, such as FRIGIDA and CaTIFY4b, emerged from our analysis. These genes not only offer insight into the molecular mechanisms underlying drought tolerance but also serve as promising targets for marker-assisted breeding, paving the way for the development of climate-resilient chickpea cultivars.
Looking Ahead
The genomic resources, detailed genetic maps, and marker-trait associations presented in our study are invaluable assets for the chickpea research community. As we move forward, we aim to translate these findings into practical breeding strategies that will enhance yield and stress adaptation in chickpea. Our team remains dedicated to pushing the boundaries of plant genomics, and we look forward to further deepening our collaboration with Dr. Varshney to transform agricultural research for a more resilient future.