Nature Biotechnology: First Domestic Goat Genome Assembly Using Optical Mapping
Our collaborative research in Nature Biotechnology presents the first high-quality reference genome of the domestic goat, leveraging innovative automated optical mapping to achieve chromosome-level assembly.
We are pleased to highlight the publication of “Sequencing and automated whole-genome optical mapping of the genome of a domestic goat (Capra hircus)” in Nature Biotechnology. This project, involving members of the XinLab team, represented a major technical leap in mammalian genomics.
Background
Goats were among the first animals domesticated by humans, providing essential resources like meat, milk, and fiber. Despite their importance, the lack of a high-quality reference genome long hindered molecular breeding and evolutionary studies. The challenge lay in the massive size and repetitive nature of mammalian genomes, which are difficult to assemble using short-read data alone.
Key Breakthroughs
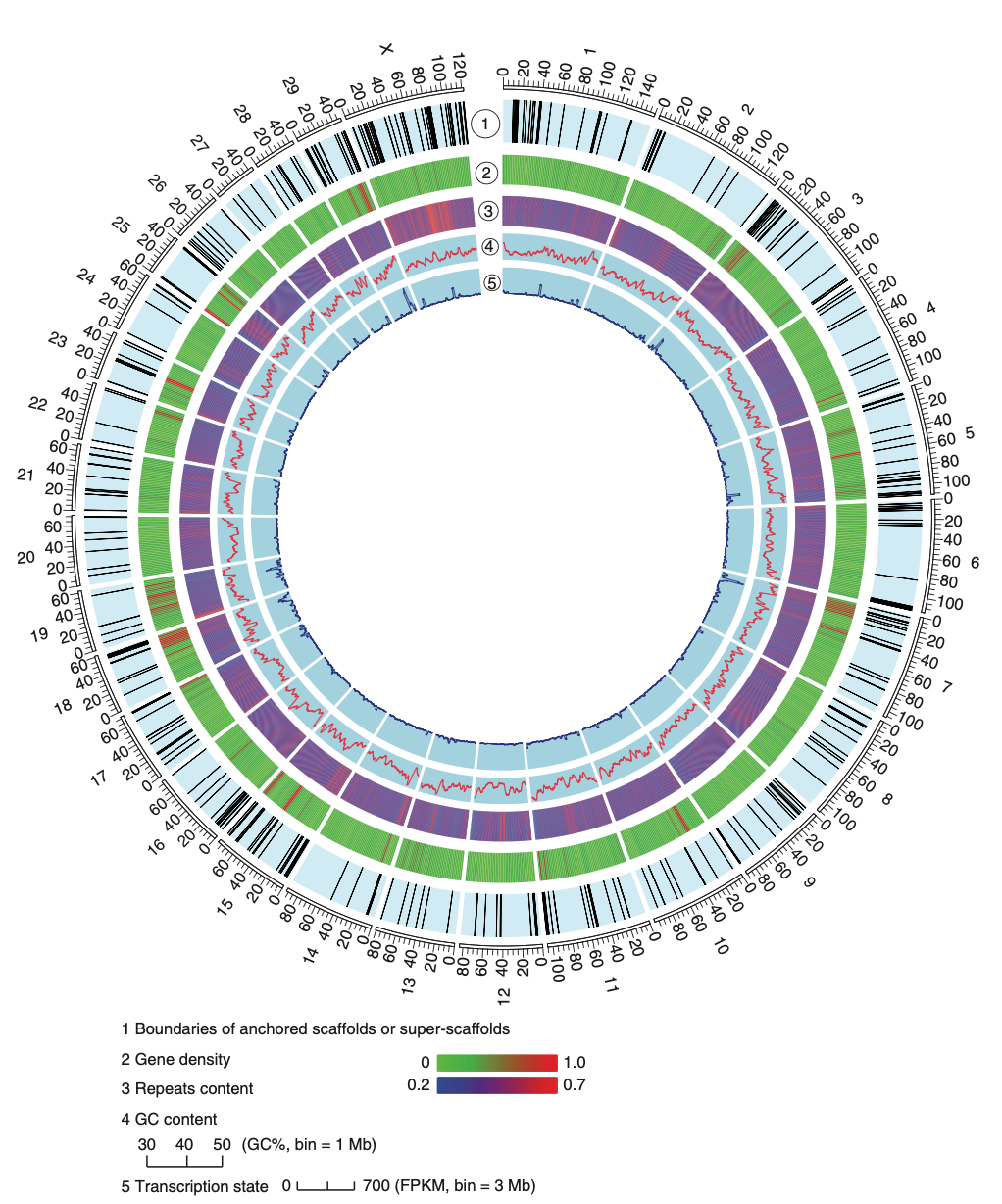
- Innovative Use of Optical Mapping: This study was one of the first to implement high-throughput whole-genome optical mapping at scale. This technology enabled the creation of super-scaffolds with an N50 of 16.3 Mb, a five-fold improvement over traditional scaffold methods, providing a robust framework for the goat’s 21 pairs of autosomes.
- Genomic Landscape: We characterized a 2.66 Gb genome containing 22,175 predicted genes. Comparative analysis highlighted gene expansions and positive selection in pathways related to immune response, digestion, and lactation, which are characteristic of ruminant biology.
- Goat vs. Sheep Divergence: By comparing the goat genome with those of sheep and cattle, we identified genetic signatures underlying the specific adaptations of goats, such as their remarkable ability to thrive in harsh environments and resist various pathogens.
Significance
The Yunnan black goat genome served as a cornerstone for the global Caprine community. It has accelerated the identification of markers for economically important traits, enabling more efficient genomic selection and precision breeding to improve yield and animal health.
Team & Collaboration
This milestone was achieved through a collaboration between the Kunming Institute of Zoology (CAS), BGI-Shenzhen, and several international partners. XinLab members (including Xin Liu and Min Xie) contributed significantly to the sequencing strategy, data integration from optical maps, and evolutionary analysis.
Read the full article here: https://doi.org/10.1038/nbt.2446